Difference between revisions of "User:Schwennesen"
Schwennesen (Talk | contribs) |
Schwennesen (Talk | contribs) |
||
| Line 242: | Line 242: | ||
And finally the two overlapping, which is specifically what I’m interested in. | And finally the two overlapping, which is specifically what I’m interested in. | ||
| − | [[ | + | [[Media:AB.png|thumb|none|alt=AB|AB]] |
I really thing that the problem is coming from the weird zig-zag on the A and tomorrow I will try to remove that from the data file to see if my results improve. Additionally, I should try to rewrite this same program using Clipper, which is the library that Dr. Puri recommended. The API seems much simpler but it doesn’t have the ability to parse from know formats like WKT or GeoJSON. I will probably also try to rewrite the same program in GEOS but using the C API (which as mentioned before can be mixed) since the C API has reentrant versions of most of the functions which will be important when using a multi-threaded application. Otherwise, I may have to add synchronization primitives like mutexes around GEOS calls which would reduce much of the concurrency. I only hope that the C API is easier to understand and has more accurate documentation then the C++ one, but that is a tomorrow problem. | I really thing that the problem is coming from the weird zig-zag on the A and tomorrow I will try to remove that from the data file to see if my results improve. Additionally, I should try to rewrite this same program using Clipper, which is the library that Dr. Puri recommended. The API seems much simpler but it doesn’t have the ability to parse from know formats like WKT or GeoJSON. I will probably also try to rewrite the same program in GEOS but using the C API (which as mentioned before can be mixed) since the C API has reentrant versions of most of the functions which will be important when using a multi-threaded application. Otherwise, I may have to add synchronization primitives like mutexes around GEOS calls which would reduce much of the concurrency. I only hope that the C API is easier to understand and has more accurate documentation then the C++ one, but that is a tomorrow problem. | ||
Revision as of 00:37, 14 June 2022
Contents
Week 1
Wednesday, 01 June 2022
This morning started with a meeting hosted by Dr. Brylow about good research practices and the importance of keeping a daily log. Both and after that, I was reading some literature that my mentor Dr. Puri had sent over. The original project that I was assigned was to use ICESAT-2 data to build a model for predicting sea ice thickness. This project was already started by a different REU student from a previous year [1] and Dr. Puri mentioned a different project, using either locality sensitive hashing and / or quantization methods to approximate nearest neighbor searches for geometry and shapes.
This new project is more appealing to me, in part because it is based on topics that I am already familiar with such as Jaccard Similarity and many notions from set theory and text mining that I have learned about at Michigan Tech. Dr. Puri sent me some preliminary literature including a chapter from what I believe is a textbook on using locality sensitive hashing to compute approximately nearest neighbors for text documents, which is a well studied problem and not the focus of this research project.
The Jaccard Similarity is a measure of the likeness between sets, but it can also be defined for areas such as polygons. For a set it is defined as
And for polygons, we do the analogous steps and take the area of the intersection or overlap and divide it by the area of the union or the total footprint of the two polygons put together.
For large datasets, we may be interested in similar objects, but for a dataset with n objects where would be C(n,2) pairs to evaluate with is at least an O(n^2) operation before adding the time needed to compute the Jaccard coefficient. The idea behind locality sensitive hashing (LSH) is to create some hash function h which is more likely to place similar items together and less likely to place dissimilar items together. Most of this information is in [2], and I should find more information about the source of this PDF that Dr. Puri sent me. He also send [3], a shorter paper that he wrote about the principles of using LSH on GIS data and some comparison to other approximate nearest neighbor (ANN) approaches.
Using the references on [3], I have found PDF copies of the remaining references below and will be reading and annotating them in the coming days. In addition to the literature review, I also need to establish the actual research question of interest here and set my sights on a realizable goal for the end of the REU. Currently, we would like to produce a small model as a proof of concept which can be demonstrated at the end of the REU.
- T. Chen, “Using Machine Learning Techniques to Analyze Sea Ice Features Using ICESAT-2 Data”.
- “Finding Similar Items”. From a currently unknown textbook.
- S. Puri, “Jaccard Similarity and LSH Applications,” ACM, Jun. 2022, doi: https://doi.org/10.1145/nnnnnnn.nnnnnnn.
- C. Fu and D. Cai, “EFANNA : An Extremely Fast Approximate Nearest Neighbor Search Algorithm Based on kNN Graph.” arXiv, Dec. 03, 2016. Accessed: Jun. 01, 2022. [Online]. Available: http://arxiv.org/abs/1609.07228
- C. Fu, C. Xiang, C. Wang, and D. Cai, “Fast approximate nearest neighbor search with the navigating spreading-out graph,” Proc. VLDB Endow., vol. 12, no. 5, pp. 461–474, Jan. 2019, doi: 10.14778/3303753.3303754.
- C. Cai and D. Lin, “Find Another Me Across the World – Large-scale Semantic Trajectory Analysis Using Spark.” arXiv, Apr. 02, 2022. Accessed: Jun. 01, 2022. [Online]. Available: http://arxiv.org/abs/2204.00878
- Y. Kalantidis and Y. Avrithis, “Locally Optimized Product Quantization for Approximate Nearest Neighbor Search,” in 2014 IEEE Conference on Computer Vision and Pattern Recognition, Jun. 2014, pp. 2329–2336. doi: 10.1109/CVPR.2014.298.
- T. Ge, K. He, Q. Ke, and J. Sun, “Optimized Product Quantization for Approximate Nearest Neighbor Search,” in 2013 IEEE Conference on Computer Vision and Pattern Recognition, Jun. 2013, pp. 2946–2953. doi: 10.1109/CVPR.2013.379.
- Y. Matsui, Y. Uchida, H. Jegou, and S. Satoh, “A Survey of Product Quantization,” vol. 6, no. 1, p. 9, 2018.
- H. Jégou, M. Douze, and C. Schmid, “Product Quantization for Nearest Neighbor Search,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 33, no. 1, pp. 117–128, Jan. 2011, doi: 10.1109/TPAMI.2010.57.
This project has the opportunity for me to learn GPU programming which is something that I’d be interested in however Dr. Puri thinks that it is not needed and most of the GPU accelerated libraries use CUDA which I cannot use since my laptop does not have an NVIDIA GPU. I could investigate HIP which is the AMD competitor to CUDA but does not have the same level of support that CUDA does. I am presently unsure if the programming portion of this project will be conducted in python or C++. These are the libraries that Dr. Puri sent me:
- Clipper (C++) A computational geometry library specializing in shape intersections.
- Shapely (Python) A computational geometry library.
- E2LSH (C++) A locality sensitive hashing library which can compute exact nearest neighbors for polygons. This would be useful as a base line to compare any approximate approaches against.
I have also identified these libraries:
- FALCONN (C++) A more recent update to E2LSH apparently.
- Scipy Spatial (Python) Part of the greater scipy package and does have limited nearest neighbor capabilities using KD-trees.
As for other pros and cons, I am more familiar with python at a whole then C++, but I am proficient in C++. Additionally, using C++ would be faster and I am be any to leverage an intermediate stage of parallelism: CPU multi-threading. This is possible in python but certainly not something that python is good at and would probably be easier then having to learn GPU programming from the ground up since have conceptually background on CPU threading. Unfortunately MTU uses a library called ThreadMentor which is not available outside of MTU department servers in any practical manner so I would have to learn the C++ standard library interface. It does look like semaphores would require C++ 20, which may or may not be supported on the department server here at Marquette that I should be getting access to in the coming days.
I an tentatively calling this project “Approximate Nearest Neighbor Searches of Real World Geometry”, but this title is subject to change once the problem becomes well defined.
Thursday, 02 June 2022
Today I continued the literature review regarding approximate nearest neighbor searches. I read three articles which I had identified yesterday:
C. Fu and D. Cai, “EFANNA : An Extremely Fast Approximate Nearest Neighbor Search Algorithm Based on kNN Graph.” arXiv, Dec. 03, 2016. Accessed: Jun. 01, 2022. [Online]. Available: http://arxiv.org/abs/1609.07228
While the graph based approaches to this do provide excellent results, the memory constraints are higher than the LSH methods used in the comparison. While the datasets used here are not as large as the ones mentioned in Dr. Puri’s LSH Applications paper, if we assume a linear memory growth, I’m not convinced that an approach like EFANNA is off the table.
C. Cai and D. Lin, “Find Another Me Across the World – Large-scale Semantic Trajectory Analysis Using Spark.” arXiv, Apr. 02, 2022. Accessed: Jun. 01, 2022. [Online]. Available: http://arxiv.org/abs/2204.00878
This paper uses minhashing, a type of LSH to find similar semantic trajectories, basically people with similar life styles based off of the places that they visit, the order of places and the amount of time spent there. They developed a hashing method called Sequence-Sensitive Hashing (SSH) which was designed to respect the sequential nature of this data. It was interesting to see LSH outside of the textbook chapter that I read about it, but the data here is still rather similar to the document structure that LSH is frequently practiced on.
T. Ge, K. He, Q. Ke, and J. Sun, “Optimized Product Quantization for Approximate Nearest Neighbor Search,” in 2013 IEEE Conference on Computer Vision and Pattern Recognition, Jun. 2013, pp. 2946–2953. doi: 10.1109/CVPR.2013.379.
This paper was somewhat of a break from the other two. Product Quantization is an approach which is adjacent to LSH and serves, at least to my current understanding, as a “different backend” for this whole process. It involves breaking the data space into a number of subspaces such that the original space is the Cartesian product of the subspaces. This produces distortion in the quantization process, which must be minimized to push for better results. The authors were able to find a non-parametric solution to this issues which can optimize the distortion in an iterative manner and it performed very well. Additional specialization is discussed for data which follows a Gaussian or normal distribution.
I additionally identified six libraries which may be relevant to this project once programming begins.
- annoy (C++ / Python) Since when did Spotify develop things like this? Thanks to the NetworkX Core Development Team for this recommendation.
- EFANNA (C++) EFANNA implementation.
- NSG (C++) NSG implementation by the EFANNA authors. This is their improvements on EFANNA I believe, but while I have the research paper for NSG I have not had the chance to read it.
- kGraph (C++ with limited Python support). ANN graph approach using a technique called NN-descent. Used as a benchmark for EFANNA.
- FLANN (C++ / Python) KD-tree based algorithms.
- yael (C++ / Python) Not sure what algorithms this uses but it was linked off the website for the datasets used in multiple of the papers I read today.
The datasets of this website where used in the Fu and Cai paper and Ge et al. papers, which may or may not be interesting resources to look at. I believe that these data sets are just numeric vectors in 128 or 960 dimensions which is idea for research on approximate nearest neighbor search (ANNS).
Looking at the programming resources that I have found, C++ is starting to pull ahead of my innate desire to program everything in python. I do not believe that this decision needs to be made this week.
One of the most important things for me to do this week is to find a narrow and tailored research topic. It looks like there as been a lot of research, which I am currently crawling through, on the general ANNS problem. However, I have not seen anything about using these techniques on shapes or other geometries. The Cai and Lin paper is probably the closest when it was discussing coordinate based similarity in their trajectories. I do need to investigate this specific sub-problem further, but I suspect that finding a suitable hashing or quantization function for the input geometries will be the heart of this problem. Once the LSH hashes or PQ codebooks are produces, I don’t see why existing methods like EFANNA or the algorithm described by Ge et al wouldn’t be applicable. However, if my experience with Google’s Summer of Code last year taught me anything it is that my last statement is almost certainly wrong and I just don’t have the knowledge yet see the problem and find a solution.
Friday, 03 June 2022
Today was another day of literature review. I read several papers, given by the below citations. I have written a few sentences about each article.
Y. Cao, T. Jiang, and T. Girke, “Accelerated similarity searching and clustering of large compound sets by geometric embedding and locality sensitive hashing,” Bioinformatics, vol. 26, no. 7, pp. 953–959, Apr. 2010, doi: 10.1093/bioinformatics/btq067.
This was a very interesting paper, and even though it was about chemical compounds, most of their work was about euclidean space which makes in intimately practical for my work with geometry which is by euclidean by definition. The interesting thing is that this work focused on a high number of dimensions, generally over 100 to create the embedding in the EI-Search and EI-Cluster algorithm. Their process picks a subset, generally three times the number of dimensions of the data to create a reference embedding then minimizes a “stress” function, basically minimizing the mean squared error of the difference between the new addition and the old reference points. Their implementation of the EI-Search algorithm can be found here and they reference using L-BFGS-B library (which scipy does have this algorithm minimized) and lshkit library which make or may not become useful.
B. Matei et al., “Rapid object indexing using locality sensitive hashing and joint 3D-signature space estimation,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 28, no. 7, pp. 1111–1126, Jul. 2006, doi: 10.1109/TPAMI.2006.148.
This paper was difficult for me to fully understand since it uses a lot of math notation that I was not familiar with, including ξ, Ξ and ζ which are not used frequently in the areas of math I am used to. It contacted several references which I did find useful, but I’m not sure how much I learned from it otherwise.
M. Astefanoaei, P. Cesaretti, P. Katsikouli, M. Goswami, and R. Sarkar, “Multi-resolution sketches and locality sensitive hashing for fast trajectory processing,” in Proceedings of the 26th ACM SIGSPATIAL International Conference on Advances in Geographic Information Systems, Seattle Washington, Nov. 2018, pp. 279–288. doi: 10.1145/3274895.3274943.
This paper was the real winner of the day. It was on the other side of Find Another Me Across the World – Large-scale Semantic Trajectory Analysis Using Spark, doing coordinate based trajectory similarities. Their algorithm places random disks on the area of interest and by tracking if the trajectories entered the circle. This hamming distance of these binary sketch of the trajectories can be used as an LSH encoding for Hausdorff distance, which is the maximum minimum distance needed to travel between two curves. By recording the order of the intersections of the circles, they produce an LSH hash family for the Frechet distance which is basically the minimum “leash” distance needed to link two points on different curves when you can only move forward on the curve. This is the more useful metric for curves. The use of randomly placed circles does remind me of the fingerprint example from the textbook chapter that I was sent. I think that trying to change this method of randomly placed circles to use the Jaccard distance, which is just 1 - Jaccard similarity, we may be able to approximate the nearest neighbor. With a fixed number and size of each circle, the preprocessing for each shape would be linear in terms of the number of circles which would be much less then the number of shapes required for the brute force all pairs solution and their data structure, the Multi-Resolution Trajectory Sketch, could be an efficient basis for a query structure. I suspect that the complexity of the Jaccard distance with the circles would be more intense then if a trajectory intersected a circle, and if so performance would never match.
G. Mori, S. Belongie, and J. Malik, “Shape contexts enable efficient retrieval of similar shapes,” in Proceedings of the 2001 IEEE Computer Society Conference on Computer Vision and Pattern Recognition. CVPR 2001, Kauai, HI, USA, 2001, vol. 1, p. I-723-I–730. doi: 10.1109/CVPR.2001.990547.
This article was an interesting read with a surprising connection to graph theory matchings that I enjoyed. However, all of their preprocessing and query operated involved at least one level of iteration over the whole dataset. For the case of the shapemes which were faster to query, the preprocessing is O(Nk) where N is the number of shape contexts (which are also expensive to compute) and k is the number of shape pieces used in the algorithm which is also an expensive operation before we get to the O(Nk) peice. This method is certainly too slow to be of use to us for this project.
It was, however, the third paper which I saw that referenced Indyk and Motwani so that will have to be the next paper that I read. A full citation is given below.
P. Indyk and R. Motwani, “Approximate nearest neighbors: towards removing the curse of dimensionality,” in Proceedings of the thirtieth annual ACM symposium on Theory of computing - STOC ’98, Dallas, Texas, United States, 1998, pp. 604–613. doi: 10.1145/276698.276876.
I also found this paper, with a new author but with the same name and 14 years more recent. I will have to find the difference and connections between these papers.
S. Har-Peled, P. Indyk, and R. Motwani, “Approximate nearest neighbors: towards removing the curse of dimensionality,” Theory of Comput., vol. 8, no. 1, pp. 321–350, 2012, doi: 10.4086/toc.2012.v008a014.
Week 2
Monday, 06 June 2022
Attended responsible conduct in research training.
Tuesday, 07 June 2022
The end is hopefully approaching for my literature review. Today I read
P. Indyk and R. Motwani, “Approximate nearest neighbors: towards removing the curse of dimensionality,” in Proceedings of the thirtieth annual ACM symposium on Theory of computing - STOC ’98, Dallas, Texas, United States, 1998, pp. 604–613. doi: 10.1145/276698.276876.
This is one of the most referenced papers that I’ve seen during the rest of my literature review, and I probably should have read it sooner. However, it seems that this paper was a big break through for approximate nearest neighbor (ANN) searches of high dimensional vectors, like other papers dedicated solely to ANN improvements have been working with. The authors use a new type of data structure called a ring-cover tree which can reduce the complexity of the ANN searches to only mildly exponential. According to the paper “For 0 < ε < 1 there is an algorithm for ε-NNS in R^d under any l_p norm which uses O(n) x O(1/ε)^d preprocessing and requires O(d) query time” [1]. This is great in and of itself, but it doesn’t translate well to my project which involves shapes and geometries. Basically, if I can reduce each shape to a vector in R^d for some constant d (which I think that I can do), then I could use methods. However, if that is the case, there are C++ libraries using more efficient algorithms like EFANNA or NSG which I could leverage without needed to implement a ring-cover tree.
S. Har-Peled, P. Indyk, and R. Motwani, “Approximate nearest neighbors: towards removing the curse of dimensionality,” Theory of Comput., vol. 8, no. 1, pp. 321–350, 2012, doi: 10.4086/toc.2012.v008a014.
Basically, [1] is a conference paper and this is the journal paper. We recently talked about the difference between the two types of papers in one of the REU lectures. A conference paper is presented at a conference and normally shorter or more preliminary results. Then, later the authors will revise the paper and contribute at least 30% more new content and them publish a journal paper which is just an expended version. All of the arguments about [1] also apply here, but the content was presented in a more concise format with some new generalizations and applications such as an approximate minimum spanning tree algorithm and extension to normed spaces rather then hamming distance discussed in the conference paper.
J. Ji, J. Li, S. Yan, Q. Tian, and B. Zhang, “Min-Max Hash for Jaccard Similarity,” in 2013 IEEE 13th International Conference on Data Mining, Dec. 2013, pp. 301–309. doi: 10.1109/ICDM.2013.119.
This paper was actually very interesting. It is a refinement on min hashing (sometimes referred to as min-wise hashing). The in the traditional min-hashing approach is to permute the two sets K times and only look at the hash value of the first element of the permutation. It can be mathematically proven that the probability of this value being the same for two sets converges to the jaccard similarity of the sets. This paper introduces a refinement to this idea called min-max hashing where instead of needing K permutations to generate a length K sketch, you only need to generate K/2 permutations and take the first and last values from the resulting re-ordered sets. Since generating and evaluating the permutations (which for practicality reasons we never fully apply to the dataset) is the most time consuming part of the min-hash algorithm, min-max hashing halves the computation time and actually improves the MSE of the approximate jaccard similarity by in effect sampling without as much replacement from the possible first elements of the permutations. In practice it seems that the variance improvements are detectable but not significant. However, the computation benefits more then justify using this technique if I end up using a min-hashing approach.
O. Chum, M. Perd’och, and J. Matas, “Geometric min-Hashing: Finding a (thick) needle in a haystack,” in 2009 IEEE Conference on Computer Vision and Pattern Recognition, Jun. 2009, pp. 17–24. doi: 10.1109/CVPR.2009.5206531.
I do not think that this paper will be relevant to my project, however some of the notions may be applicable and you never know what will become important. This paper is using images and the idea of visual words which I believe are simple geometric patterns that can be identified with a feature extractor like a specific curve or likewise. Note that a feature from the feature detector can contain multiple visual words. The author’s use primary visual words and features who are composed of only one visual word and a set of secondary visual words, visual words which are selected in the general proximity of the primary visual word with some restrictions on the distance and sizes allowed. Once the signatures are generated in this manner the rest of the process is very similar to the traditional min-hashing approach, but the more careful generation of the signatures proved very helpful in improving the results of the traditional min-hashing algorithm.
I have identified one more C++ library. This one is CGAL and it does have the capabilities of reading the input format that the datasets that I have identified will be presented in (more on that below). More exploration is needed to see the full extent of it’s support.
Also, I met with Dr. Puri today and learning several things. First, the textbook chapter that he sent me has the full citation presented below:
- J. Leskovec, A. Rajaraman, and J. D. Ullman, Mining of Massive Datasets, 2nd ed. [Online]. Available: http://infolab.stanford.edu/~ullman/mmds/book.pdf
There is also a third edition of this book, which is listed as having new information about min-hashing:
- J. Leskovec, A. Rajaraman, and J. D. Ullman, Mining of Massive Datasets, 3rd ed. [Online]. Available: http://infolab.stanford.edu/~ullman/mmds/book0n.pdf
During that same meeting with Dr. Puri we also discussed several technical components for the project. First, I can get datasets from UCR Star, a repository of real-world spatial datasets represented in the well-known text format. We decided that for the time being we do not need to consider the rotation of the shapes, i.e. if I rotate a square 45 degrees it is now considered a different shape when it was before. This should save a lot of time during the preprocessing steps, but accuracy of the model would certainly be improved if we could detect and rotate the shapes as needed. The only method for overcoming this that comes to mind immediately is to find the longest angle bisector in the polygon (quick StackOverflow link in case I have time for this.) and try to align it to be either vertical or horizontal in hopes that it would cause very similar shapes to be detected.
Additionally, Dr. Puri doesn’t seem concerned about needed to translate the shapes to be centered on the origin, but I am. Intuitively the jaccard similarity is the intersecting area over the union of the two shapes. If the shapes are not touching or overlapping at all then the similarity will always be zero (and thus the distance variant always one) even if it is the same shape that has been translated in a random direction. It is possible that the computational geometry libraries that I will leverage are able to account for that, but I’m not sure if they will. If not, it will be necessary to translate the shape so that the centroid, sometimes called a barycenter, is at the origin. According to the wiki page linked previously, if can find the centroid of a polygon with an O(n) operation where n is the number of vertices in the polygon. I have actually already had to implement this for Altas, a class project at Michigan Tech and I remember that something with signed area of zero was very problematic, so I hope that I don’t have that issue this summer. (link here) I think that is it unlikely since we were using a lot of artificially generated geometry in that project and not real world data like in this case.
It turns out that this problem was inspired by a problem that the National Geospatial-Intelligence Agency published as a project. This project was actually to use images from social media which have been stripped of metadata to find the nearest neighbor (or approximate nearest neighbor) in a database with tens of terabytes of geo-tagged images to help identify where the image was taken, which would be useful in both helping first responders in disaster sites or locate other individuals of interest to the government. While that first one is absolutely beneficial, the second use certainly has some ethic consideration. This project may be inspired by that work, but we are not using images of any kind in this project. Information in the NGA grant can be found below.
- “BIG-R BAA Topic 4: High Dimensional Nearest Neighbor.” Mar. 15, 2021. [Online]. Available: https://www.grants.gov/web/grants/search-grants.html?keywords=HM0476-20-BAA-0001
Dr. Puri also recommended looking into spatial hashing and z-order curves which are methods for taking discrete x and y coordinates and transforming them into one value that can be used in a traditional one dimensional hash table. I’m not sure if or when I will need to use these at the moment, but I did find two papers on spatial hashing which I will probably read tomorrow.
- S. Lefebvre and H. Hoppe, “Perfect spatial hashing,” ACM Trans. Graph., vol. 25, no. 3, pp. 579–588, Jul. 2006, doi: 10.1145/1141911.1141926.
- E. J. Hastings, J. Mesit, and R. K. Guha, “Optimization of Large-Scale, Real-Time Simulations by Spatial Hashing,” Proc. 2005 Summer Computer Simulation Conference, vol. 37, p. 9, Jul. 2005, [Online]. Available: https://www.eecs.ucf.edu/~jmesit/publications/scsc%202005.pdf
I also got login credentials to everest, a sever run by Marquette University to conduct my research on. I am planning on making a concurrent program to help move through the datasets I will be working on (generally gigabytes in size with millions of records). I would like to use the C++ standard library for my threading. That was introduced in C++11, and counting_semaphore was not introduced until C++20 so I had to determine which C++ standards were available on everest. Using the below script, I found that complying without the -std flag defaults to C++98 and that up to C++11 is supported. Since counting_semaphore is not among these, if I need can either use monitors or use a monitor to create my use semaphore. From my experience in concurrent, I will probably choose the latter. On the off chance that the C++ threads are limited to only one core (which I think is very unlikely) I will have to move to POSIX threading using pthreads.
#include <iostream>
using namespace std;
int main()
{
if (__cplusplus == 202002L) cout << "C++20" << endl;
else if (__cplusplus == 201703L) cout << "C++17" << endl;
else if (__cplusplus == 201402L) cout << "C++14" << endl;
else if (__cplusplus == 201103L) cout << "C++11" << endl;
else if (__cplusplus == 199711L) cout << "C++98" << endl;
else cout << "pre-standard C++" << endl;
return 0;
}
Wednesday, 08 June 2022
More research papers! Today I read the below papers:
B. R. Vatti, “A generic solution to polygon clipping,” Commun. ACM, vol. 35, no. 7, pp. 56–63, Jul. 1992, doi: 10.1145/129902.129906.
This is the original research paper for the general polygon clipping algorithm used as part of the clipper library which I will need to compute the jaccard distance for the shapes in my datasets. Technically, since I am planning on using a library for this, I did not need to read the paper. However, a lot of time in ANN search research is spent on the complexities of the data structure and processes so a generally understanding of the algorithm it to my direct benefit. The algorithm itself sweeps the overlapping polygons from bottom to top and divides edges into two categories; contributing or non-contributing. Contributing edges effect the final result of the clipped polygons while non-contributing ones do not. The paper lies up several rules which can be used to determine which is which. According to the paper, the total complexity of the algorithm is linear with respect to the total number of edges between the target and clipping polygon.
C. Fu, C. Xiang, C. Wang, and D. Cai, “Fast approximate nearest neighbor search with the navigating spreading-out graph,” Proc. VLDB Endow., vol. 12, no. 5, pp. 461–474, Jan. 2019, doi: 10.14778/3303753.3303754.
This was another paper about ANN, or more accurately AKNN. It builds on the EFANNA paper to further improve on four aspects which the authors have deemed important for a graph based ANN approach:
- Guarantee that the graph is connected, so that the query is always reachable.
- Reduce the average out-degree of the graph which improves search time.
- Shorten the path from the starting node to the query, which also improves search time.
- Reduce the memory usage of the graph to improve scalability.
To accomplish these goals, the authors adapted the idea of a relative neighborhood graph which is a graph on a set of points for which the longest side of each possible triangle on the set of points. However this is actually a too selective process and can lead to non-monotonic paths in the graph, i.e. paths were the only way to get closer to the query point is to temporarily move away from it. They then develop the idea of a monotonic relative neighborhood graph (MRNG) where certain edges that wouldn’t be included in the RNG are added back. The naive implementation of this O(n^2 log n + n^2 c) where c is the the average out-degree of the graph, which is clearly unacceptable for large datasets. However, it is possible to approximate the MRNG with the navigating spreading-out graph (NSG) which starts with the nearest neighbor graph (or an approximation of it, like from EFANNA) which only ensures a monotonic path from one starting location, chosen to be the mediod of the data, to all other points in the dataset. This approach leads to the fastest index building times and the smallest index which makes NSG the known graph based method. While it’s memory usage is still higher then some LSH methods, the authors were able to test it on a 100 million point data set on a machine with 96 Gb of ram successfully, so it should work for my datasets if applicable. That last bit is the trick here… Is a possible to adapt this for what I need? The original algorithm is defined only for the L2 norm do if I can use that distance measure at some point, I should be ok. If a need to use something like Manhattan distance or something more exotic I may have more problems.
Thursday, 09 June 2022
I finally did it! A day of research which did not involve be reading research papers for 6 to 8 hours!
So, if I wasn’t reading all day, what did I do?
- Create the daily log from yesterday (I know, I just forgot before going to bed last night).
- Fleshed out the stub page for my REU project, attempting to find research goals and summarize the problem. Recall that the project is called Approximate Nearest Neighbor Searches of Real World Geometry.
- Started trying to formulate the plans and theoretical properties that I am going to need to prove the correctness of whatever system I develop and explore different methods that I could follow. That document is typeset in LaTeX given the higher mathematics dense then I could reasonably support in this wiki (please give us MathJax or KaTeX on the wiki) and I have emailed it to Dr. Puri to get his feedback on it.
- It seems like
everestdoes not have all of the dependencies for some of the libraries that other researchers have developed so I also inquired into what process will be needed to install those. If they cannot be installed there are work-arounds but they will be slower and I will not have time nor the ability to rewrite these libraries at the same level of optimization.
Friday, 10 June 2022
- Y. Liu, J. Yang, and S. Puri, “Hierarchical Filter and Refinement System Over Large Polygonal Datasets on CPU-GPU,” in 2019 IEEE 26th International Conference on High Performance Computing, Data, and Analytics (HiPC), Dec. 2019, pp. 141–151. doi: 10.1109/HiPC.2019.00027.
This was another paper sent to me by Dr. Puri and it is about using a hierarchical filtering system to perform operations like spatial join over large geospaital datasets. To do this they developed the notion of a PolySketch which is based off breaking the polygon or line into segments then find the minimum bounding rectangles. Finding the intersections of the rectangles is easier then of the polygons themselves due to the simpler geometry. The usage of minimum bounding rectangles is more traditional within the land of computer graphics and very easy to work with. This is however different from “Multi-resolution sketches and locality sensitive hashing for fast trajectory processing” by Astefanoaei et al which has been one of my important references since they use circles. The circles are an important choice since offsetting them in any direction will produce the same area profile which they use build the probabilistic bounds needed for proving the LSH family’s properties. Because of this, the use of circles here is going to be similarly important to this work. Unfortunately I do not believe that Clipper has dedicated circle support which means that I will have to use an approximation but of course the computational complexity of computing the Jaccard distance varies linearly with the total number of vertices between both so the better the approximation the longer the operations will be. This will likely become a user setting.
In addition, I began to consider some of the more technical details of the project. I am currently thinking that I will start testing with the lakes dataset on UCR Star. This set is 7.4 GB and has 8 million records and can be downloaded in
- GeoJSON
- CSV
- KMZ
- Shapefile
- JSON+WKT
This means that I have a choice about how I want to parse this data. Python has much better support for these data formats than C++ does (shocker, I know), so there will likely be a preprocessing step done in python. Please note that for all of these ANN searches, the time to build the index is critical to evaluating the method, so I cannot do too much with this. I have downloaded a sample of the lakes dataset in both JSON+WKT and CSV and both of them contain the WKT data, just embedded in different data storage formats for the auxiliary data like the name of the body of water. I am currently not sure if this auxiliary data will need to be preserved in the final data structure, but it may be useful. Python does have a JSON library and CSV is not overly difficult to parse through, but I plan to use Clipper for Jaccard distance which does not read WKT so I will most likely parse each WKT representation to a text file where each point is a new line, which would be very easy to read in C++.
Another possible preprocessing data step might be to center each of the polygons around it’s centroid so that the step doesn’t need to be repeated while working on only the data structures. However, this is an important part of the creation of the data structure indices, so the option to perform it in the C++ portion of the code should at least exist.
The data is very large and I would like to preprocess concurrently so first I’d split the file into manageable chunks. On linux systems, sed -n '$=' <filename> can find the number of lines in a file and split -l <number> <filename> will split the file into chucks with the given number of lines. By combining these I can create the correct number of new files programmatically based on the number of processes that will be run.
Monday, 13 June 2022
Today I worked on setting up my development environment on everest. I think that using some of the libraries that I have discovered I can develop this application excursively in C++ using tools like
- GEOS - the computational geometry library that shapely wraps. Note that the C++ API is unstable, but I don’t expect to be updating the library during the course of this summer. Additionally, it is possible to run C code in C++ and the C API for GEOS is stable. GEOS has WTK and GeoJSON readers. Another advantage for the C interface is that their are reentrant versions of most of the functions.
- gnuplot - command line plotting library that I found a C++ wrapper for. I actually used gnuplot in high school for a few weeks plotting paths for the robotics team I was a part of.
And I started by installing gnuplot locally on everest, which was not that difficult to build from source. The tricky part was the C++ integration with gnuplot. This is a single C++ header file which uses POSIX pipes to connect to the gnuplot binary which I put unto my PATH. Since I am running only an ssh connection to everest, it does not connect to the X server and naturally the DISPLAY environment variable is left undefined. This is fine since I can set gnuplot to output a png, jpeg or a whole host of other file types instead of displaying an X window. The C++ header file that I found even includes a static method called set_terminal_std to set this value. However, I was constantly getting an exception saying that the DISPLAY environment variable could not be found. After trying every imaginable combination of where and what to call set_terminal_std with I finally opened the header file to track down the problem, which I had determined was coming from the constructor for the gnuplot object using valgrind. At the top the init function used by all of the various constructors I found this (starting at line 1694):
#if ( defined(unix) || defined(__unix) || defined(__unix__) ) && !defined(__APPLE__)
if (getenv("DISPLAY") == NULL)
{
valid = false;
throw GnuplotException("Can't find DISPLAY variable");
}
#endif
So, even if I was already using the static method set_terminal_std to change the terminal type to output a png, it was always still going to error. Looking at the source code for set_terminal_std (which was included in the doxygen documentation that I had generated) I was able to correct this line to the below code so that if the terminal type was x11, the default value and the DISPLAY variable couldn’t be found it would generate the exception.
#if ( defined(unix) || defined(__unix) || defined(__unix__) ) && !defined(__APPLE__)
if (terminal_std.find("x11") != std::string::npos && getenv("DISPLAY") == NULL)
{
valid = false;
throw GnuplotException("Can't find DISPLAY variable");
}
#endif
And suddenly everything was working properly.
Getting GEOS to work was also very difficult. In order to even install it I had to download a more recent version of cmake and configure my PATH to find that before the version already on everest. But in the scheme of things, that was easy compared to what came next. I decided to write a small program using GEOS which read the large polygons that Dr. Puri had sent. The GEOS documentation was not very helpful. While they did warn that the C++ API was unstable, I thought that it would be fine since I would only need to work with the current version. However, there were type alaises and functions which
- The most recently documentation includes
- StackOverflow posts from almost 10 years ago include
- The header files themselves include, but yes I got to the point where I was reading them
But g++ couldn’t find them. Eventually, after about 5 hours I was able to read the shapes and find the function call to perform an intersection of the shapes (since that is what this library will be doing for me, in additional to reading the geometry from the data files) to be meant with an error message about “non-noded intersections” which I definitely did not understand. Google searches showed that many other programmers using computational geometry libraries were encountering this, but the term “non-noded geometries” was not formally defined anywhere and explanations about what exactly it is varied but included self-intersecting polygon and line segments which were sitting too close to each other.
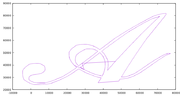
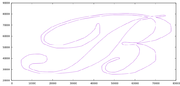
So I decided that I needed to see the shapes to understand if anything was wrong. Them in gnuplot revealed what I believe is the problem.
And finally the two overlapping, which is specifically what I’m interested in.
I really thing that the problem is coming from the weird zig-zag on the A and tomorrow I will try to remove that from the data file to see if my results improve. Additionally, I should try to rewrite this same program using Clipper, which is the library that Dr. Puri recommended. The API seems much simpler but it doesn’t have the ability to parse from know formats like WKT or GeoJSON. I will probably also try to rewrite the same program in GEOS but using the C API (which as mentioned before can be mixed) since the C API has reentrant versions of most of the functions which will be important when using a multi-threaded application. Otherwise, I may have to add synchronization primitives like mutexes around GEOS calls which would reduce much of the concurrency. I only hope that the C API is easier to understand and has more accurate documentation then the C++ one, but that is a tomorrow problem.